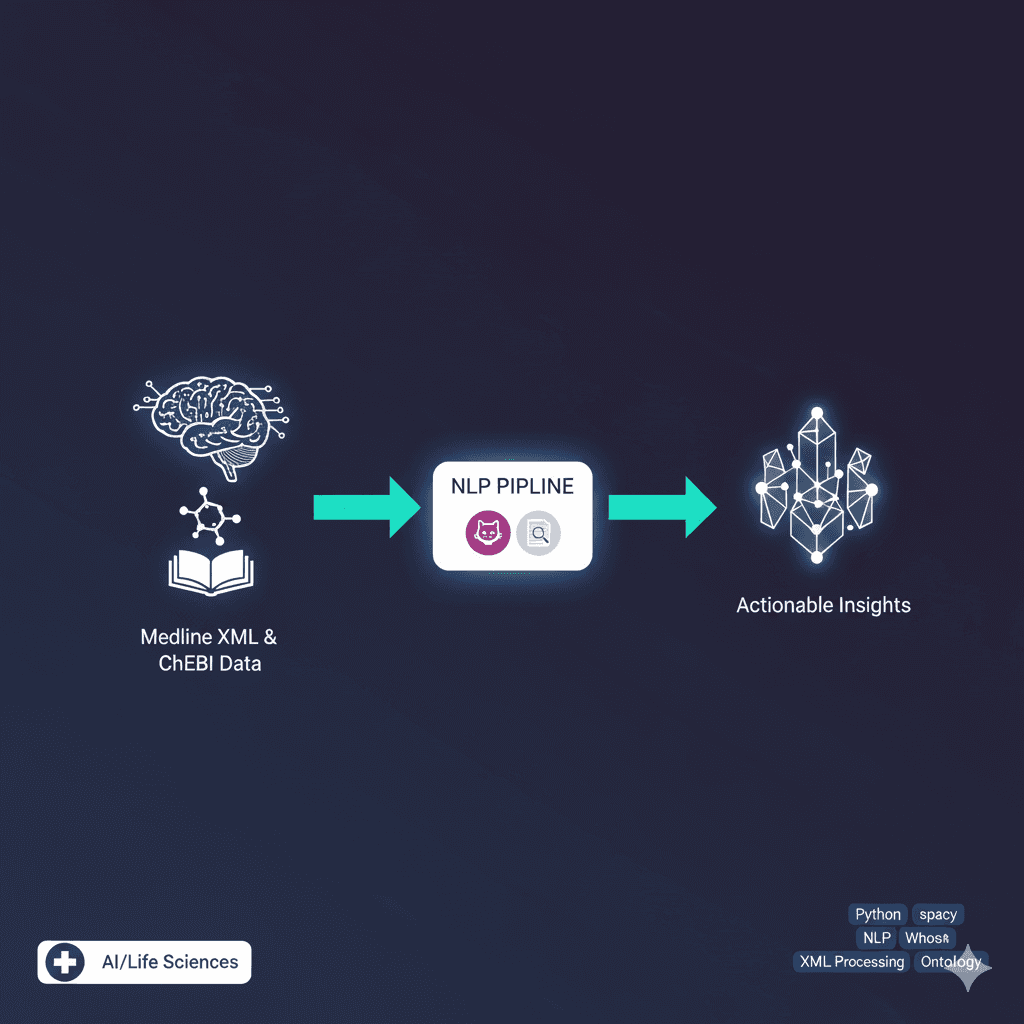
Medical Knowledge Graph Construction with MedCAT
Developed a comprehensive medical knowledge graph system using MedCAT for entity extraction and Neo4j for storage, integrating SNOMED CT medical terminology.

Project Overview
Built a medical knowledge graph construction pipeline as part of my Master's studies in computational life sciences. The system uses MedCAT (Medical Concept Annotation Toolkit) for automated medical entity extraction from clinical texts and integrates SNOMED CT medical terminology for standardized concept mapping. The knowledge graph is stored and queried using Neo4j graph database with RDF framework for managing medical relationships. This project provided hands-on experience with medical NLP, healthcare ontologies, and graph database technologies. I learned about the challenges of processing medical text data, entity linking, and building semantic relationships between medical concepts. The work demonstrates practical applications of knowledge graphs in healthcare and clinical decision support systems.
Key Features
- ✓MedCAT medical entity extraction
- ✓SNOMED CT terminology integration
- ✓Neo4j graph database implementation
- ✓RDF framework for semantic relationships
- ✓Clinical text processing pipeline
- ✓Medical concept annotation and mapping
- ✓Standardized healthcare terminology
- ✓Graph-based medical knowledge representation
- ✓Efficient entity relationship storage
- ✓Medical ontology integration
- ✓Clinical decision support capabilities
- ✓Scalable healthcare data architecture
Technical Challenges
- ⚡Integrating complex medical terminology standards
- ⚡Optimizing Neo4j performance for medical data
- ⚡Ensuring accurate medical entity recognition
- ⚡Managing SNOMED CT concept hierarchies
- ⚡Handling clinical text variations and ambiguity
- ⚡Designing efficient graph schema for medical relationships
- ⚡Maintaining data privacy for healthcare information
- ⚡Scaling entity extraction for large clinical datasets
Technologies Used
Project Info
Screenshots